Jump to: Untargeted Lipidomics | Experimental Design | Fatty Acid Methyl Esterification (FAME) | Short Chain Fatty Acid Characterization (SCFA)
Untargeted Lipidomics
This page describes unbiased acquisition of relative quantification of lipid species spanning a large range of lipid classes.
Experimental design
Untargeted lipidomics experiments are designed to acquire data on many individual lipid species and compare abundances between experimental conditions (e.g., drug vs DMSO, knockout vs. wild type, etc.). Comparison between conditions requires a minimum of three biological replicates per condition.
Ideally enough sample material is provided to create a pooled quality control sample (by combining equal volume from each sample). A powerful way to validate identified lipid features is by comparing relative abundance of a potential lipid feature between a quality control and diluted (typically two-fold) quality control samples. Features that do not have a change in abundance consistent with the known dilution are excluded.
The pooled quality control sample is also used to monitor instrument variability throughout the course of a LC-MS queue, with increasing importance as experiment times/number of samples increase.
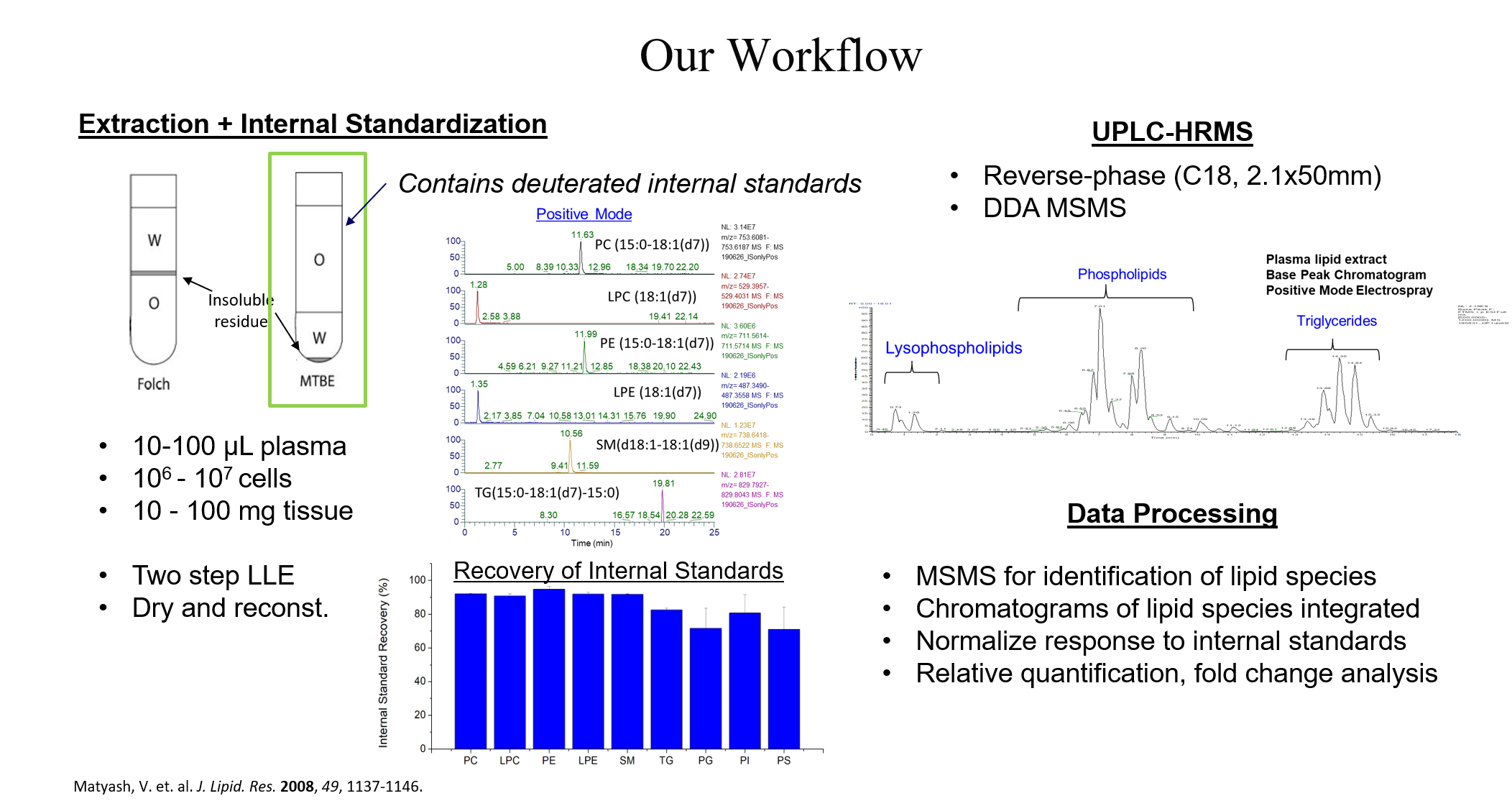
Untargeted lipidomics is accomplished using liquid-liquid extraction of raw biological material (biofluids, cells, tissue, etc.) followed by liquid chromatography coupled to a high-resolution mass spectrometer using data-dependent acquisition of tandem mass spectra (DDA).
Lipids are separated by hydrophobicity using reverse-phase liquid chromatography and introduced to the MS using electrospray ionization (ESI). DDA provides a combination of high resolution, accurate mass data at the MS1 level (which can suggest molecular formulas) and structural information obtained at the MS2 level (which can suggest connectivity within a molecule) that can be used to identify lipid species.
Sample specific information
- Biofluids (plasma, cerebrospinal fluid, bronchoalveolar lavage, etc.)
- Easiest to work with by liquid-liquid extraction method
- Volumes between 10-200 μL (higher volumes are needed for lipid-deficient fluids and vice-versa)
- Generally able to add internal standards (in methanol) directly for protein precipitation
- Cells
- Preferably suspended in PBS and flash frozen
- 1×106 is a good starting point for most cell types, decreasing counts will decrease number of detectable lipids
- Can add internal standards (in methanol) directly for lysis and protein precipitation
- Tissue
- Likely need to be homogenized prior to analysis
- 1-10 mg
- Add PBS to homogenized tissue prior to addition of internal standards